Multiscale imaging methods
Integrative approach
Understanding a process from the molecular to the cellular scale and eventually to the level of the microbial community in an environmental sample requires an approach that reveals insights at different scales of resolution. Cryo-electron tomography (cryoET) bridges Structural Biology with Cell Biology and resolves complexes in their cellular context. We integrate cryoET imaging data with single-particle cryoEM structure determination and (time-lapse) light microscopy. The multiscale imaging approach is further complemented by biochemical experiments, proteomics, genomics, functional assays and microbiological methods.
Method development
Driven by our biological questions, we develop methods that enable:
- improved cryoET data acquisition.
- improved and automated sample preparation by focused ion beam milling.
- imaging of samples with higher complexity and size.
- identification of cellular structures and cells in cryo-tomograms.
CryoET data acquisition
Recent advances in instrumentation and software allowed us to significantly external page speed-up data collection and were the basis for the development of external page PACE-tomo and external page SPACE-tomo by our colleague and former lab member Fabian Eisenstein.
Sample thinning by cryo-focused ion beam milling
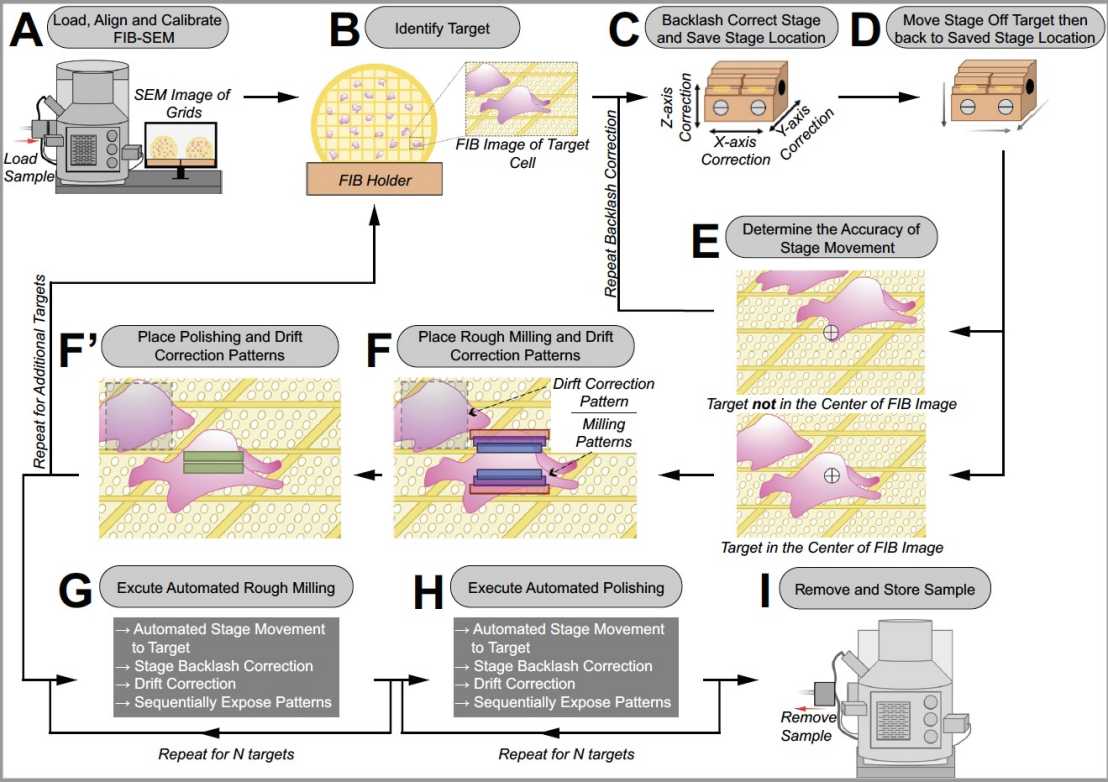
Since cryoET is limited to relatively thin samples, most cellular samples have to be thinned prior to imaging. We have established a external page workflow using cryo-focused ion beam milling to generate lamellae of different organisms. In collaboration with Zeiss, we also developed a method for external page automating the sequential generation of lamellae, which our lab and many other groups now routinely apply in their research.
We are currently engaged in developing workflows that enable the thinning of samples with larger volumes by plasma-focused ion beam milling using a ThermoFisher Arctis instrument.
Identification of filaments in cryo-tomograms
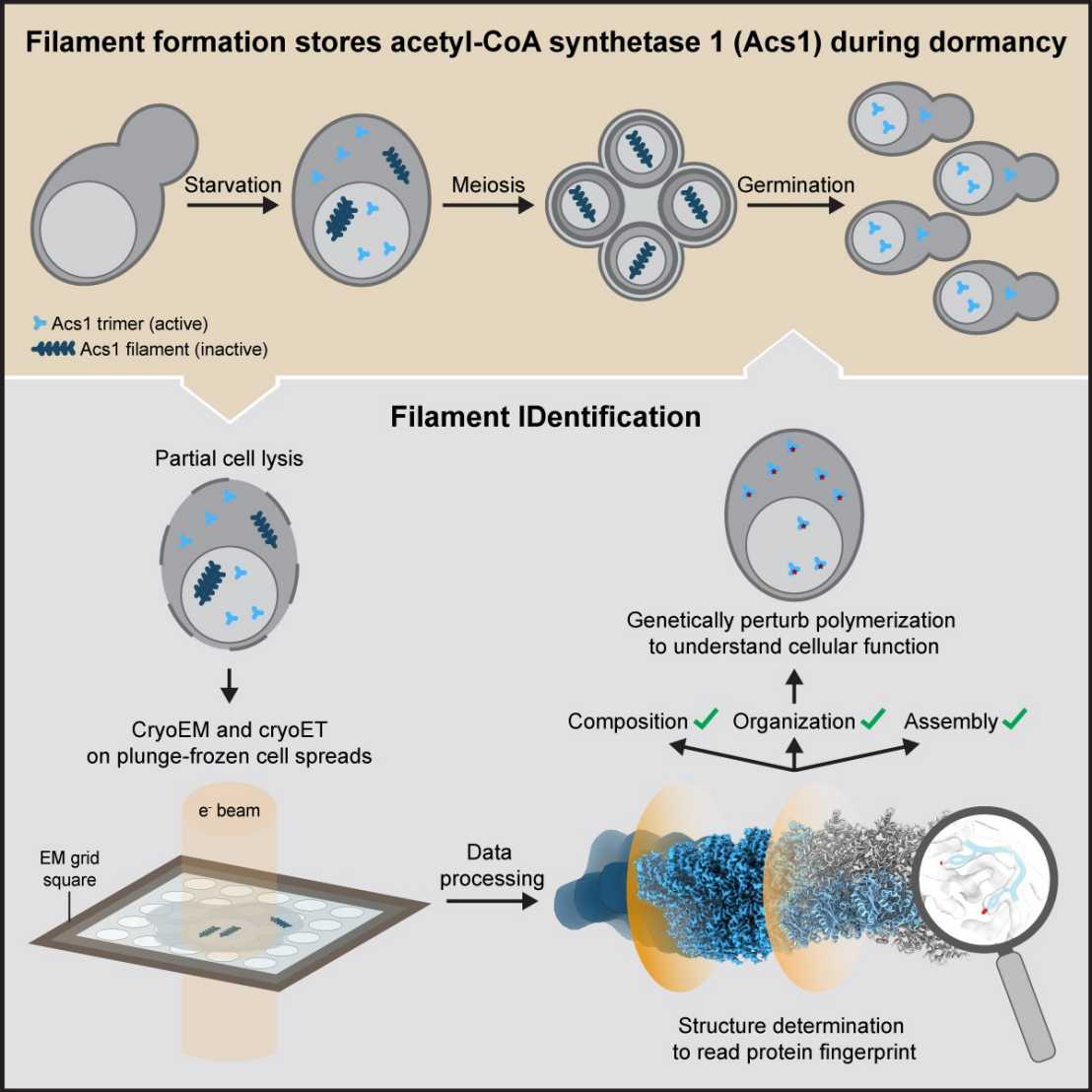
By cryoET imaging, we often discover intriguing cellular structures with unknown identity. We have developed external page FilamentID in order to uncover the previously unknown identity of filaments of metabolic enzymes in different cellular compartments of yeast. The approach is based on a combination of cryoET and single-particle cryoEM of gently lysed and spread cells.
external page In bacteria, we identified cytoplasmic filaments consisting of a protease by a combination of cryoET and subtomogram averaging, proteomics and structure prediction methods.
Identification of cells in cryo-tomograms
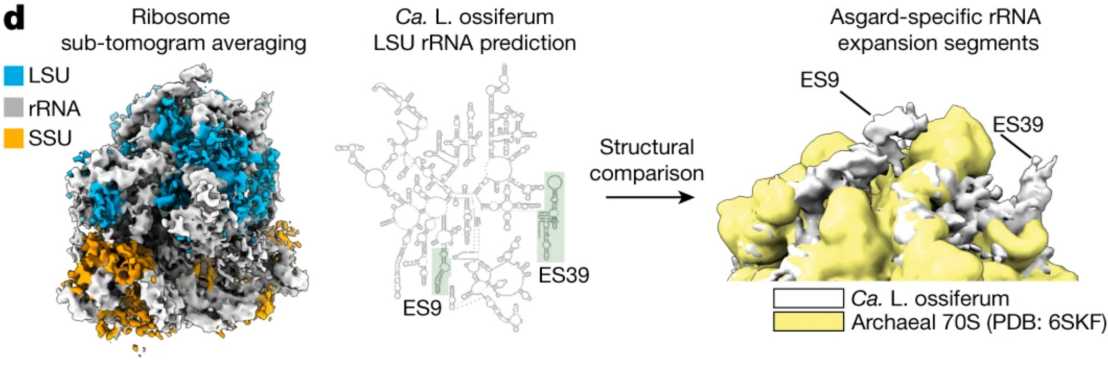
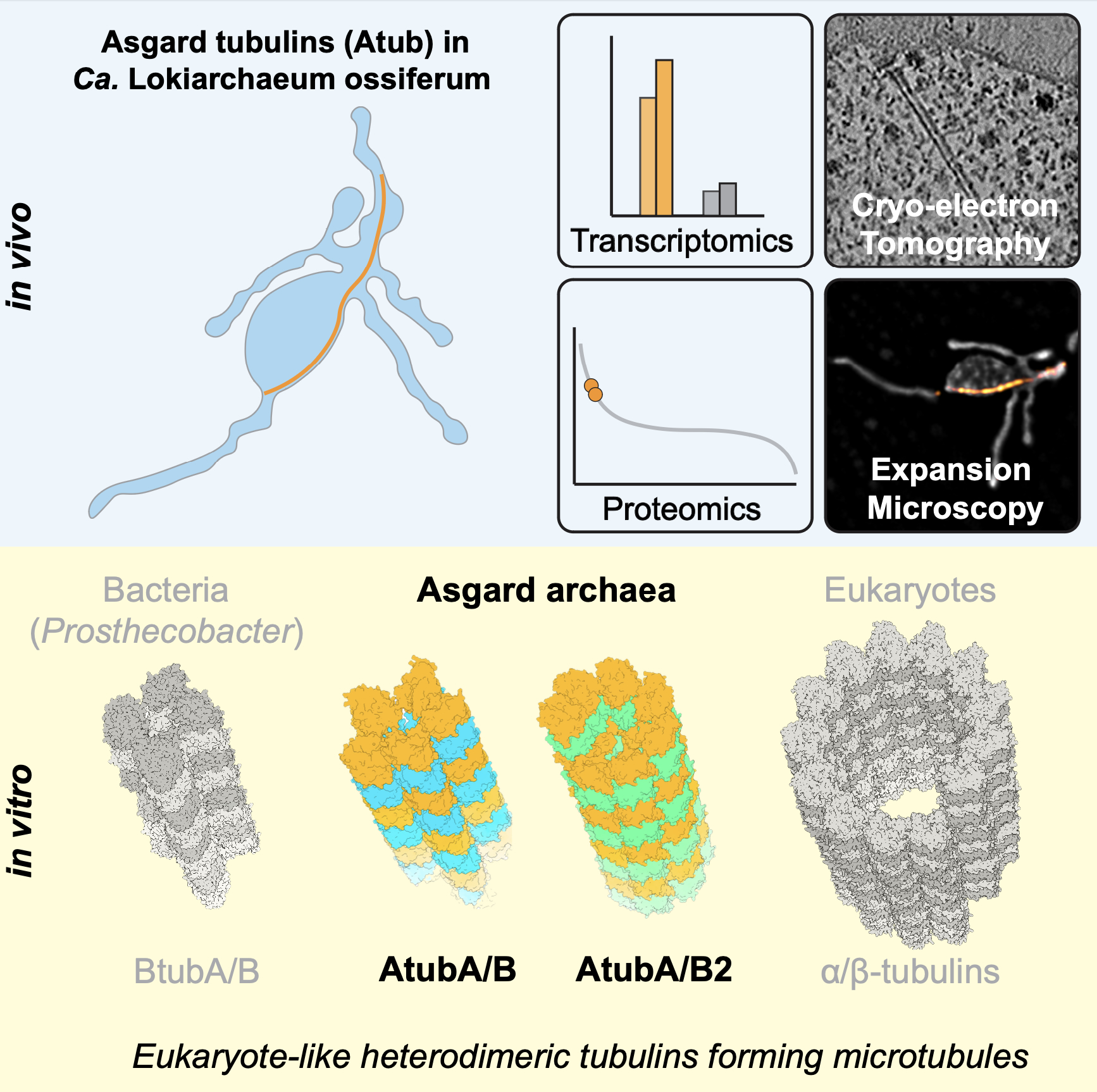
When imaging a non-pure culture by cryoET, we sometimes face the challenge of identifying and distinguishing cells in such a complex sample. In the case of Asgard archaea, external page we tackled this problem by generating a subtomogram average of ribosomes that allowed for a structure-based identification of Asgard cells.
We are now working on methods that can more generally be applied to identify cells in complex environmental or patient samples.